Abstract
Programmed cell death ligand 1 (PD-L1) plays a key role in tumor immune escape by negatively regulating cytotoxic T-cells (CTLs) via PD-1 receptors. Thus, those tumors with high PD-L1 expression are thought to be sensitive to PD-1/PD-L1 blockade, reactivating CTL reactions to tumors. However, the mechanism that regulates PD-L1 expression in tumor cells has not been fully elucidated, understanding of which would help to develop effective anti-tumor immunotherapy.
Recently, we reported that disruption of 3'-UTR of PD-L1 led to a remarkably enhanced expression of PD-L1 in a wide variety of human cancers, particularly adult T-cell leukemia/lymphoma (ATL) and diffuse large B-cell lymphoma. In these cancers, stability of PD-L1 transcripts is negatively regulated via their 3'-UTR sequence, whose disruption thus, results in markedly elevated PD-L1 expression and immune evasion (Kataoka et al., Nature, 2016). It has been well established that non-coding regions (i.e., 5'- and 3'-untranslated regions (UTRs)) play important roles in the regulation of mRNA expression, which is accomplished by several transacting factors that bind to cis-regulatory elements within the UTR. Based on this knowledge, we investigated the mechanism that controls PD-L1 expression through 3'-UTR sequence, primarily focusing on those transacting RNA-binding proteins (RNA-BPs).
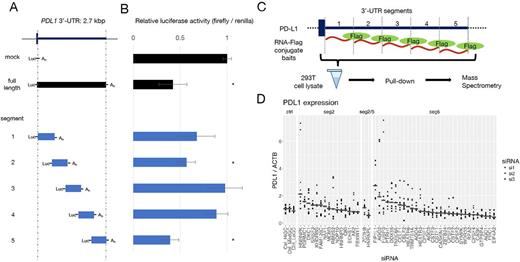
To determine the relevant regions within the 3'-UTR which are capable of repressing PD-L1, several cell lines were transfected with luciferase reporter vectors, in which a luciferase coding sequence was concatenated to various deletion mutants of the PD-L1 3'-UTR (Panel A). We identified two critical sequences, segment 5 and 2 within the PD-L1 3'-UTR, whose existence significantly decreased luciferase expression (Panel B; 293T data, mean ± SD, * denotes t-test p < .05). Importantly, the deletion of these sequences showed a similar effect on gene expression in various cell lines derived from different tissues. To confirm this finding, we used a CRISPR-mediated tiled 3'-UTR editing in situ technique. We designed all possible single-guide (sg) RNAs in the PD-L1 3'-UTR. The library was virally infected into cells and those with high expression of PD-L1 were concentrated by FACS, and the enrichment of each sgRNA was evaluated by high-throughput sequencer. The enriched sgRNAs localized to positions compatible with the luciferase assay, confirming that the two regions are actually responsible for the regulation of PD-L1 expression.
Next, we searched RNA-BPs that bind to these regions in mass spectrometry, utilizing flag-peptide-tagged RNA pull-down method (Panel C). In brief, we synthesized PD-L1 3'-UTR RNA segments along with β-actin mRNA as control in vitro, and conjugated a flag-peptide to their 3'-ends. Using these RNA-baits, an immunoprecipitation experiment was performed in 293T cells and the co-immunoprecipitated proteins were analyzed by mass spectrometry. The data obtained from different segments were compared to each other. In addition to those proteins binding to multiple regions within the PD-L1 3'-UTR, we found proteins that specifically interacted with the repressive segments (segment 5 and 2) identified through luciferase assays.
Finally, to confirm that the proteins detected by mass spectrometry actually suppress PD-L1 expression, we performed knock-down experiments using siRNA designed for the RNA-BPs that are presumed to interact with the segment 5 and 2. Three siRNA constructs per gene were transfected to 293T cells and their effect on PD-L1 expression was evaluated by RQ-PCR. Panel D shows relative PD-L1 expressions for each siRNA targets with median line, which are grouped according to relevant PD-L1 3'-UTR segments. The negative regulatory effect of these RNA-PBs on PD-L1 expression was largely confirmed. PD-L1 protein level was also increased, when these genes were knocked out by CRISPR/Cas9 system. Expression of these genes in ATL and other lymphomas was also evaluated.
In summary, we identified critical sequences within the PD-L1 3'-UTR and RNA-BPs that bind to these sequences and negatively regulate PD-L1 expression. Our findings should not only provide novel insight into the molecular mechanisms by which PD-L1 expression in tumor cell is regulated but also help to identify potential targets for immune therapy.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.